deLemus
Dynamic Expedition of Leading Mutations in SARS-CoV-2 Spike Glycoproteins
News

The dynamic epidemiology of coronavirus disease 2019 (COVID-19) since its outbreak has been a result of the continuous evolution of its etiological agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Within the first 2 years of this pandemic, the World Health Organization (WHO) has already announced five variants of concern (VOC), namely Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529), together with numerous variants of interest (VOI).[1] The emergence of all these variants has brought along many novel mutations that continue to fine-tune the fitness of the virus, leading to its persistent global circulation.[2],[3]
The purpose of deLemus platform is to continuously monitor the evolutionary trajectories of SARS-CoV-2 spike glycoproteins. By employing low-rank tensor decomposition and epistatic correlation analysis on reported sequences, we provide the list of outlined leading mutations on a monthly basis.[4] The outlined leading mutations unravel the evolutionary mutation pattern of the spike glycoprotein, establishing the deLemus platform as an early warning system for upcoming variants and facilitating timely contributions in the ongoing efforts to combat SARS-CoV-2.
Spike Glycoprotein
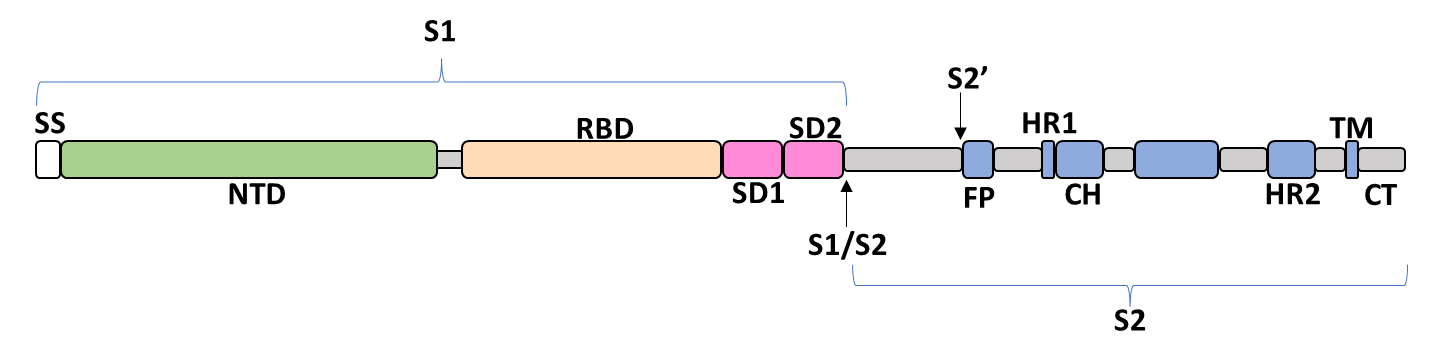
The spike glycoprotein of SARS-CoV-2 is a trimeric type I viral fusion protein that binds the virus to the angiotensin-converting enzyme 2 (ACE2) receptor of a host cell.[5] It is composed of 2 subunits: the N-terminal subunit 1 (S1) and C-terminal subunit 2 (S2), within which multiple domains lie. The S1 region facilitates ACE2 binding and is made up of an N-terminal domain (NTD), a receptor-binding domain (RBD), and 2 C-terminal subdomains (CTD1 and CTD2), while the downstream S2 region is responsible for mediating virus-host cell membrane fusion.
References
- Karim, S. S. A. & Karim, Q. A. Omicron SARS-CoV-2 variant: A new chapter in the COVID-19 pandemic. Lancet 398, 2126 (2021).
- Carabelli, A. M. et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat Rev Microbiol 21, 162 (2023).
- Witte, L. et al. Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. Nat Commun 14, 302 (2023).
- Kolda, T. G. & Bader, B. W. Tensor Review. SIAM Rev 51, 455 (2009).
- Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23, 3 (2021).
- Cov-Lineages. https://cov-lineages.org/
- GISAID. https://gisaid.org/